Single-cell RNA sequencing (scRNA-seq) resolves cellular heterogeneity beyond population averages. Current workflows enable novel cell discovery and cellular dynamics analysis. Future expansion depends on shifting from transcriptional profiling to integrated, multi-dimensional biological modeling using multiomics and spatial transcriptomics, while overcoming challenges in standardization, noise control, and computational scaling.
Current scRNA-seq focuses on transcriptomes. Future pipelines will integrate DNA, ChIP, ATAC, and epigenetic modifications with RNA. This enables direct analysis of regulatory mechanisms controlling gene expression, not just expression patterns. Integrated omics will improve understanding of cell fate regulation and phenotypic control.
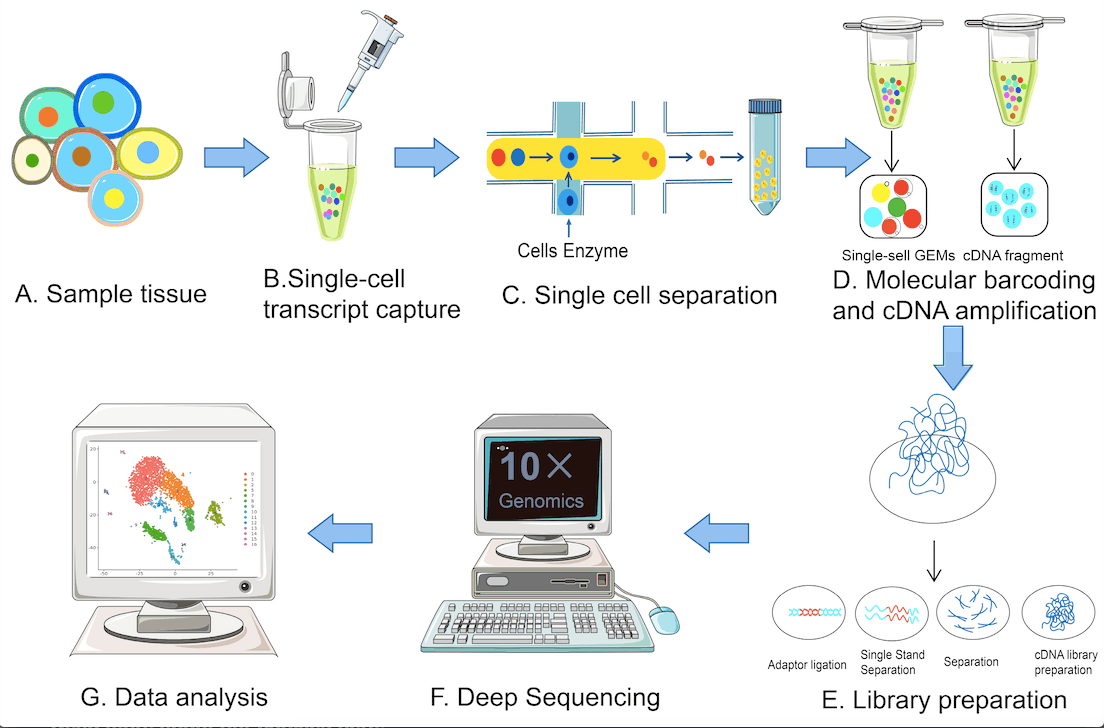
The single-cell sequencing techniques typically involve the following steps: single-cell isolation, reverse transcription, cDNA synthesis, single-cell library construction, high-throughput sequencing, and data analysis. The following figure illustrates the various steps.
Standard scRNA-seq loses spatial information during tissue dissociation. Spatial transcriptomics restores cell position within tissues, enabling analysis of tissue architecture, cellular neighborhoods, and spatiotemporal interactions. This is critical for heterogeneous systems such as organoids and improves reconstruction of functional tissue organization.
scRNA-seq is advancing toward patient-specific high-throughput screening for personalized therapy. This requires machine-learning–driven clinical pipelines capable of handling complex patient data.
Future analysis shifts from static clustering to dynamic trajectory inference using pseudotime to model biological processes. RNA velocity adds directional prediction, allowing estimation of future cellular states.
Over 600 standalone tools exist without universal standards. Steps such as normalization, clustering, and integration vary between platforms (Seurat vs. Scanpy; R vs. Python). This causes inconsistent outputs from the same dataset and limits cross-study reproducibility.
scRNA-seq data is zero-inflated due to the dropout effect, with only 1–5% transcript capture per cell. This creates low signal-to-noise ratios and high variability. Ambient RNA, lysed cells, and empty droplets introduce further artifacts. Accurate interpretation requires robust quality-control filtering, including Dirichlet-multinomial models for droplet discrimination.
Datasets now scale from thousands to millions of cells, causing memory overload and curse-of-dimensionality errors in traditional tools. Expansion depends on high-scaling platforms (e.g., Scanpy) and efficient dimensionality reduction (e.g., UMAP) without data loss.
Summary of Key Factors
| Category | Key Elements | Impact |
|---|---|---|
| Emerging Tech | Multiomics | Integrates DNA, RNA, epigenetics |
| Spatial Transcriptomics | Restores tissue structure | |
| Trajectory Inference | Models dynamic biological processes | |
| Analytical Challenges | Standardization | Inconsistent results across tools |
| Data Sparsity | Dropout obscures true biology | |
| Scalability | Tools must handle >1 million cells |
The future of scRNA-seq depends on integrating multiomics and spatial transcriptomics to capture full biological complexity. Simultaneously, standardized, scalable, and noise-robust computational pipelines are essential to ensure reproducibility, accuracy, and clinical applicability.
“Future pipelines must cope with multiomics data integration.” — Slovin et al.