RNA interference (RNAi) is a conserved process that regulates gene expression using small silencing RNAs acting in a sequence-specific manner. Fire and Mello first described this phenomenon in C. elegans in 1998. Earlier, researchers called it post-transcriptional gene silencing (PTGS), transgene silencing, or quelling. RNAi occurs in all eukaryotes—from yeast to mammals—and plays crucial roles in regulating gene expression, controlling transposons, and protecting cells against viral infections. The two main classes of small silencing RNAs involved are microRNAs (miRNAs) and small interfering RNAs (siRNAs).
miRNAs in RNAi
MiRNAs are small non-coding RNAs encoded by the genomes of plants, animals, and their viruses. These molecules, typically 20-25 nucleotides long, regulate gene expression by binding to the 3′-untranslated regions (3′-UTR) of specific messenger RNAs (mRNAs). The first miRNA, lin-4, was discovered by Victor Ambros and his team in C. elegans. Unlike protein-coding genes, lin-4 produces a short RNA complementary to target mRNAs, preventing gene translation. Later in 2000, Gary Ruvkun’s group discovered another miRNA, let-7, which showed similar gene-regulating activity.
MiRNAs influence developmental timing, cell differentiation, proliferation, cell death, and oncogenesis. Their biogenesis begins in the nucleus when RNA polymerase II transcribes a long primary transcript (pri-miRNA). Next, the RNase III enzyme Drosha, along with DGCR8 in mammals (or Pasha in Drosophila), processes pri-miRNAs into 70-100 nucleotide hairpin structures called pre-miRNAs. Exportin-5 then transports these pre-miRNAs to the cytoplasm for further processing.
In the cytoplasm, the enzyme Dicer cleaves the pre-miRNAs into mature miRNA duplexes. These duplexes load into the RNA-induced silencing complex (RISC). Argonaute (Ago) proteins play a critical role within RISC by facilitating gene silencing. Specifically, the PAZ domain of Ago interacts with the miRNA’s 3′ end, while its PIWI domain, similar to ribonuclease-H, executes the silencing function.
Differences in miRNA Biogenesis Between Plants and Animals
Plant miRNA biogenesis differs from that in animals. Plants rely on a single enzyme, DCL1, which performs the functions of both Drosha and Dicer. Moreover, before exiting the nucleus, plant miRNA duplexes undergo methylation by the enzyme HEN1. This modification does not occur in animal miRNAs.
miRNAs and Gene Silencing Mechanisms
MiRNAs silence genes either by degrading target mRNAs or inhibiting their translation. This outcome depends on how closely the miRNA complements its target mRNA. High complementarity usually results in mRNA cleavage by Ago proteins, while partial complementarity inhibits translation. Typically, miRNAs recognize targets using their “seed sequence,” a crucial 2-8 nucleotide stretch at the miRNA’s 5′ end.
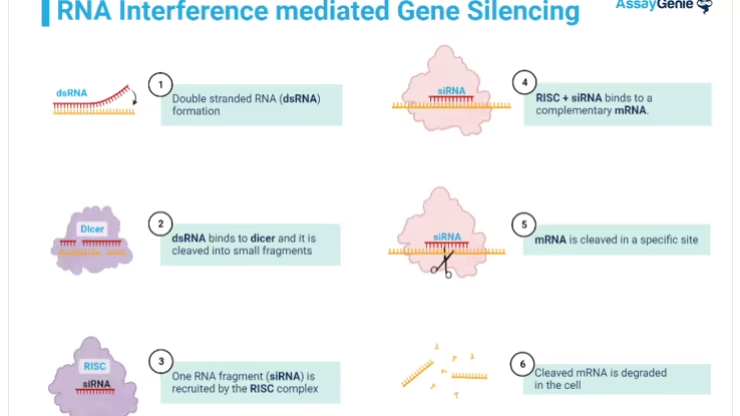
RNAi Mediated by siRNAs
In the siRNA pathway, double-stranded RNAs (dsRNAs) undergo processing into siRNA duplexes by the Dicer enzyme. These siRNAs are approximately 21 nucleotides long, featuring two-nucleotide 3′ overhangs. Dicer, a large multi-domain RNase III enzyme (~200 kDa), cleaves dsRNA into siRNAs and packages them with a dsRNA-binding protein into RISC.
Within RISC, the guide strand of siRNA hybridizes with the target mRNA through full complementarity, allowing RISC to silence the gene. Meanwhile, the passenger (or anti-guide) strand degrades during activation. Argonaute proteins act as endonucleases, cleaving the target mRNA precisely at the complementary site.
Conclusion: Key Differences Between miRNAs and siRNAs
The primary difference between siRNAs and miRNAs lies in their origins. SiRNAs derive from double-stranded RNA precursors, whereas miRNAs come from single, extended transcripts that fold into imperfect hairpin structures. Typically, siRNAs exhibit full complementarity to their targets, causing precise cleavage. Conversely, miRNAs usually show partial complementarity, which leads to translational repression of multiple mRNAs sharing similar sequences.
piRNAs: A New Class of Small RNAs
More recently, scientists discovered piRNAs, small RNAs about 25-30 nucleotides long that bind Piwi clade Argonaute proteins. Unlike siRNAs and miRNAs, piRNAs originate from single-stranded RNA precursors and do not require Dicer processing. Most piRNAs are antisense but can be sense as well and feature 2′-O-methylation at their 3′ ends, similar to siRNAs. PiRNAs may suppress transposon expression by either cleaving mRNA or modifying chromatin states at transposon loci.
Engage with Us
Stay tuned for more captivating insights and News. Visit our Blogs , Science paper , Study Portal and Follow Us on social media to never miss an update. Together, let’s unravel the mysteries of the natural world.